Add full name and description for target genes
Add full name and description for target genes
.
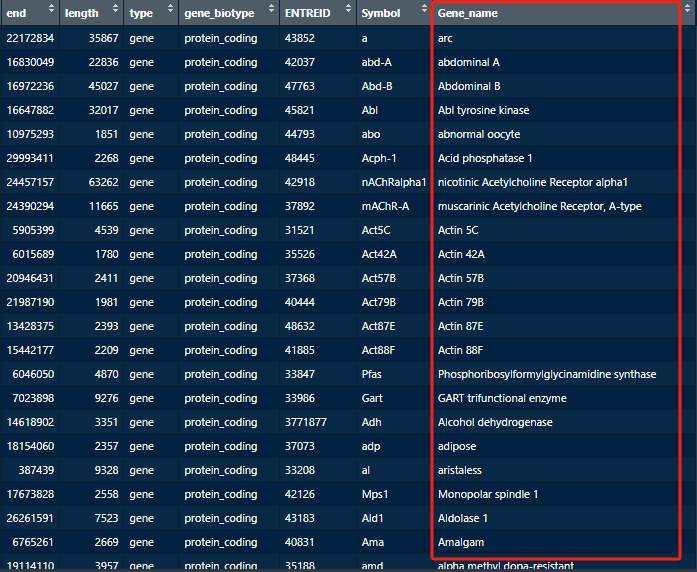
Some genes’ functions may not be easily inferred from their abbreviated names, therefore, to be better understand the genes, we need to include their full names and other forms of ID numbers for the target genes.
1 | |
output file :
the full names are indicated by the red box.
Add full name and description for target genes
https://www.lianganmin.cn/2025/01/10/Add-Full-Name-and-Description-for-Target-Gene/