1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
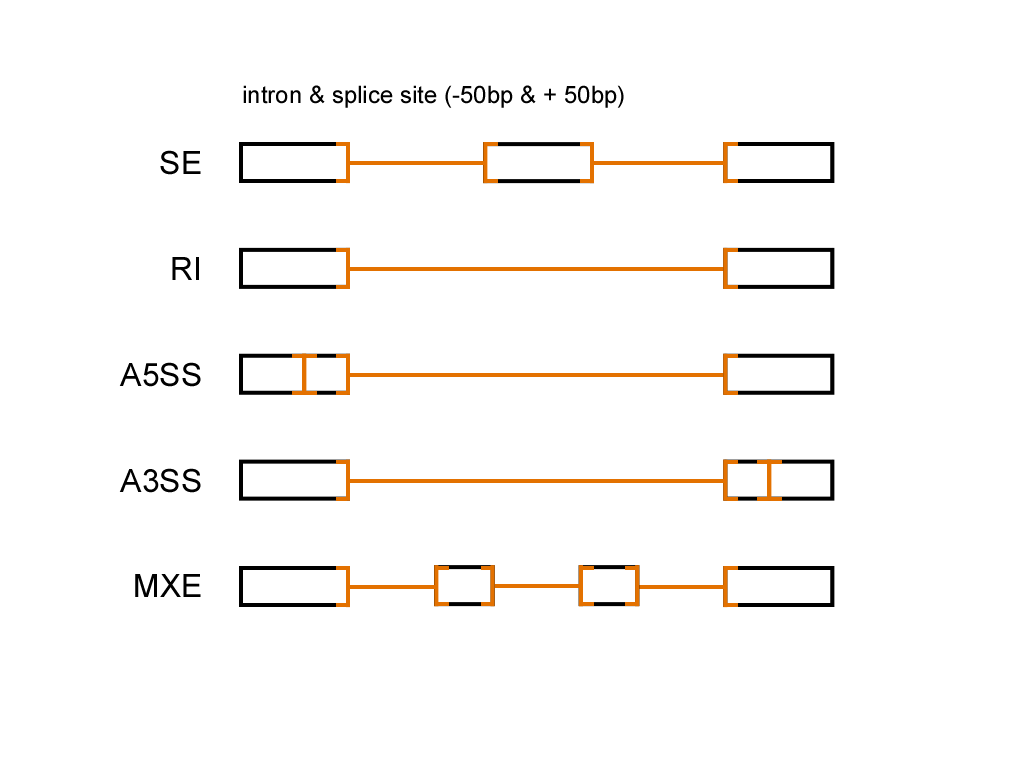
| # ------ splice site
# SE
setwd("E:/220625_PC/R workplace/220320_SXL/220421_SXL.S2/AS/230806_AS/raw_gtf/novel/")
se_sxl_novelSS_S2 <- read.table("SE.MATS.JC.txt",header = T)
se_sxl_novelSS_S2 <- merge(se_sxl_novelSS_S2,genome.anno, by="GeneID", all.x=TRUE)
se_sxl_novelSS_S2$chr <- gsub("chr", "", se_sxl_novelSS_S2$chr)
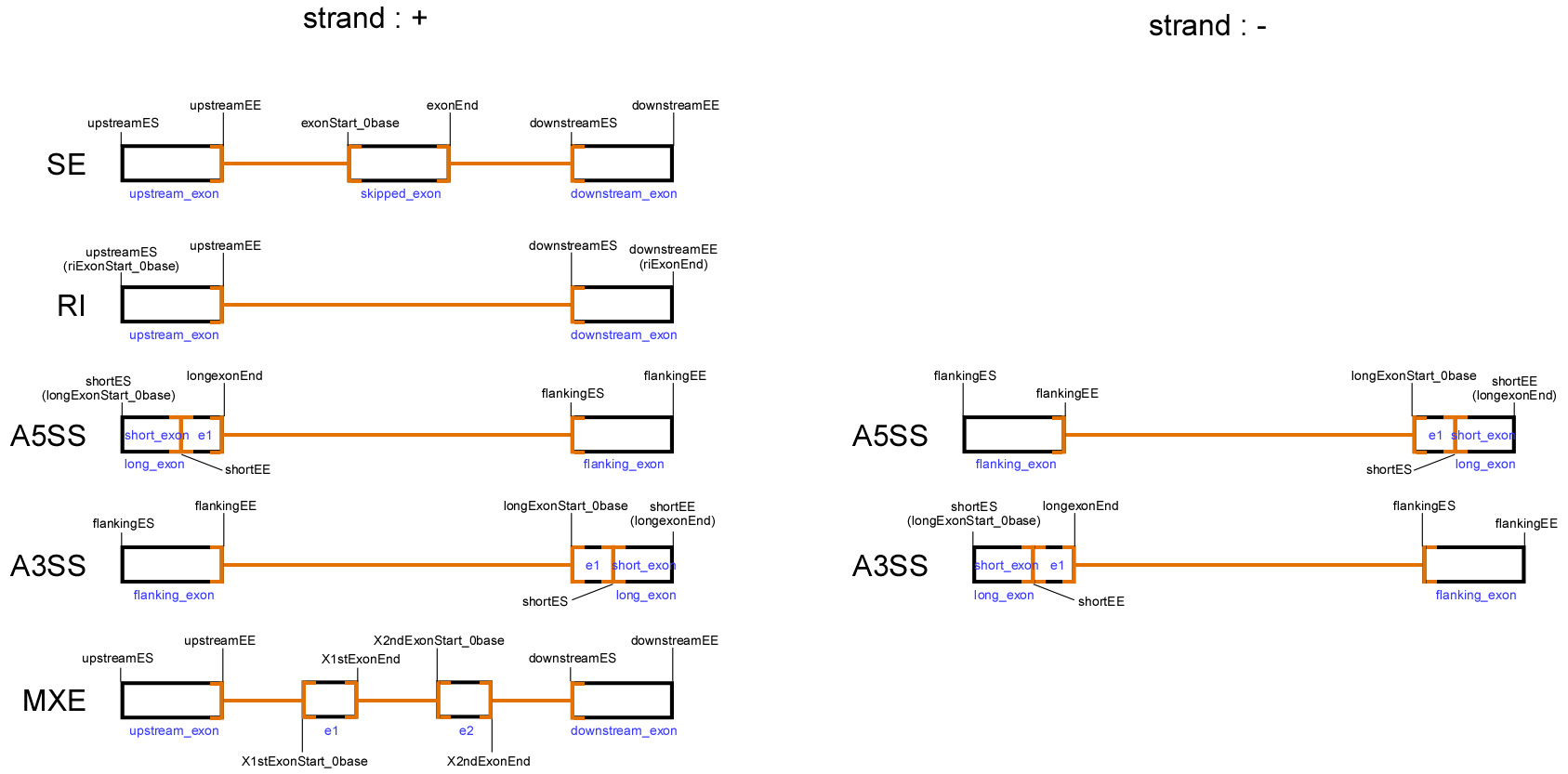
S2_se_upstreamEE <- se_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","upstreamEE")]
S2_se_exonStart_0base <- se_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","exonStart_0base")]
S2_se_exonEnd <- se_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","exonEnd")]
S2_se_downstreamES <- se_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","downstreamES")]
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/250318/splice.site/")
write.table(S2_se_upstreamEE,file = "S2_se_upstreamEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_se_exonStart_0base,file = "S2_se_exonStart_0base.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_se_exonEnd,file = "S2_se_exonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_se_downstreamES,file = "S2_se_downstreamES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
# RI
setwd("E:/220625_PC/R workplace/220320_SXL/220421_SXL.S2/AS/230806_AS/new/without_novel/")
ri_sxl_novelSS_S2 <- read.table("RI.MATS.JC.txt",header = T)
ri_sxl_novelSS_S2 <- merge(ri_sxl_novelSS_S2,genome.anno, by="GeneID", all.x=TRUE)
ri_sxl_novelSS_S2$chr <- gsub("chr", "", ri_sxl_novelSS_S2$chr)
S2_ri_upstreamEE <- ri_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","upstreamEE")]
S2_ri_downstreamES <- ri_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","downstreamES")]
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/250318/splice.site/")
write.table(S2_ri_upstreamEE,file = "S2_ri_upstreamEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_ri_downstreamES,file = "S2_ri_downstreamES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
# A5SS
setwd("E:/220625_PC/R workplace/220320_SXL/220421_SXL.S2/AS/230806_AS/raw_gtf/novel/")
a5ss_sxl_novelSS_S2 <- read.table("A5SS.MATS.JC.txt",header = T)
a5ss_sxl_novelSS_S2 <- merge(a5ss_sxl_novelSS_S2,genome.anno, by="GeneID", all.x=TRUE)
a5ss_sxl_novelSS_S2$chr <- gsub("chr", "", a5ss_sxl_novelSS_S2$chr)
a5ss_sxl_novelSS_S2_p <- a5ss_sxl_novelSS_S2[a5ss_sxl_novelSS_S2$strand == "+",]
a5ss_sxl_novelSS_S2_m <- a5ss_sxl_novelSS_S2[a5ss_sxl_novelSS_S2$strand == "-",]
S2_a5ss_p_shortEE <- a5ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","shortEE")]
S2_a5ss_p_longExonEnd <- a5ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","longExonEnd")]
S2_a5ss_p_flankingES <- a5ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","flankingES")]
S2_a5ss_m_flankingEE <- a5ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","flankingEE")]
S2_a5ss_m_longExonStart_0base <- a5ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","longExonStart_0base")]
S2_a5ss_m_shortES <- a5ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","shortES")]
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/250318/splice.site/")
write.table(S2_a5ss_p_shortEE,file = "S2_a5ss_p_shortEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a5ss_p_longExonEnd,file = "S2_a5ss_p_longExonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a5ss_p_flankingES,file = "S2_a5ss_p_flankingES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a5ss_m_flankingEE,file = "S2_a5ss_m_flankingEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a5ss_m_longExonStart_0base,file = "S2_a5ss_m_longExonStart_0base.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a5ss_m_shortES,file = "S2_a5ss_m_shortES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
# A3SS
setwd("E:/220625_PC/R workplace/220320_SXL/220421_SXL.S2/AS/230806_AS/raw_gtf/novel/")
a3ss_sxl_novelSS_S2 <- read.table("A3SS.MATS.JC.txt",header = T)
a3ss_sxl_novelSS_S2 <- merge(a3ss_sxl_novelSS_S2,genome.anno, by="GeneID", all.x=TRUE)
a3ss_sxl_novelSS_S2$chr <- gsub("chr", "", a3ss_sxl_novelSS_S2$chr)
a3ss_sxl_novelSS_S2_p <- a3ss_sxl_novelSS_S2[a3ss_sxl_novelSS_S2$strand == "+",]
a3ss_sxl_novelSS_S2_m <- a3ss_sxl_novelSS_S2[a3ss_sxl_novelSS_S2$strand == "-",]
S2_a3ss_p_flankingEE <- a3ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","flankingEE")]
S2_a3ss_p_longExonStart_0base <- a3ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","longExonStart_0base")]
S2_a3ss_p_shortES <- a3ss_sxl_novelSS_S2_p[,c("GeneID","ID","gene_symbol","chr","strand","shortES")]
S2_a3ss_m_shortEE <- a3ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","shortEE")]
S2_a3ss_m_longExonEnd <- a3ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","longExonEnd")]
S2_a3ss_m_flankingES <- a3ss_sxl_novelSS_S2_m[,c("GeneID","ID","gene_symbol","chr","strand","flankingES")]
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/250318/splice.site/")
write.table(S2_a3ss_p_flankingEE,file = "S2_a3ss_p_flankingEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a3ss_p_longExonStart_0base,file = "S2_a3ss_p_longExonStart_0base.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a3ss_p_shortES,file = "S2_a3ss_p_shortES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a3ss_m_shortEE,file = "S2_a3ss_m_shortEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a3ss_m_longExonEnd,file = "S2_a3ss_m_longExonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_a3ss_m_flankingES,file = "S2_a3ss_m_flankingES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
# MXE
setwd("E:/220625_PC/R workplace/220320_SXL/220421_SXL.S2/AS/230806_AS/raw_gtf/novel/")
mxe_sxl_novelSS_S2 <- read.table("MXE.MATS.JC.txt",header = T)
mxe_sxl_novelSS_S2 <- merge(mxe_sxl_novelSS_S2,genome.anno, by="GeneID", all.x=TRUE)
mxe_sxl_novelSS_S2$chr <- gsub("chr", "", mxe_sxl_novelSS_S2$chr)
S2_mxe_upstreamEE <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","upstreamEE")]
S2_mxe_X1stExonStart_0base <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","X1stExonStart_0base")]
S2_mxe_X1stExonEnd <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","X1stExonEnd")]
S2_mxe_X2ndExonStart_0base <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","X2ndExonStart_0base")]
S2_mxe_X2ndExonEnd <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","X2ndExonEnd")]
S2_mxe_downstreamES <- mxe_sxl_novelSS_S2[,c("GeneID","ID","gene_symbol","chr","strand","downstreamES")]
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/250318/splice.site/")
write.table(S2_mxe_upstreamEE,file = "S2_mxe_upstreamEE.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_X1stExonStart_0base,file = "S2_mxe_X1stExonStart_0base.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_X1stExonEnd,file = "S2_mxe_X1stExonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_X2ndExonStart_0base,file = "S2_mxe_X2ndExonStart_0base.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_X2ndExonEnd,file = "S2_mxe_X2ndExonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_X2ndExonEnd,file = "S2_mxe_X2ndExonEnd.txt",quote = F,row.names = F,col.names = F,sep = '\t')
write.table(S2_mxe_downstreamES,file = "S2_mxe_downstreamES.txt",quote = F,row.names = F,col.names = F,sep = '\t')
|