1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
|
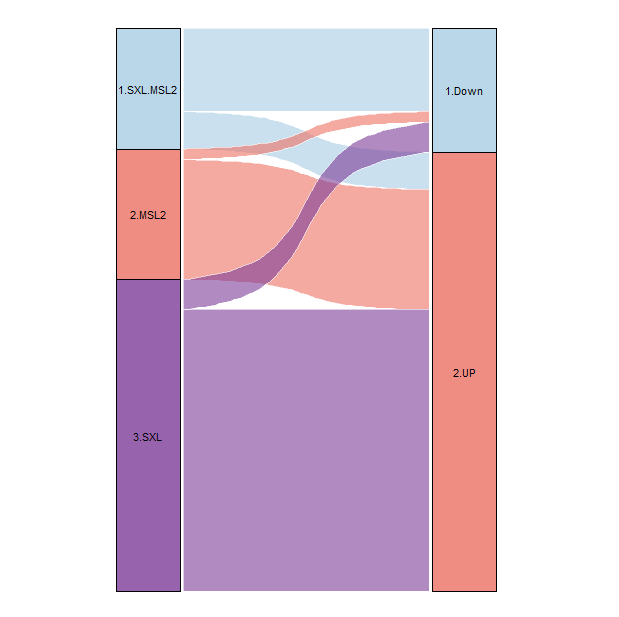
S2_DEG <- rbind(S2_up[,c(1,3)],S2_down[,c(1,3)])
S2_DEG$S2.TYPE <- ifelse(S2_DEG$GeneID %in% msl2_in_genes_gene$geneID &
S2_DEG$GeneID %in% peak_SXL_genes$GeneID, "1.SXL.MSL2",
ifelse(S2_DEG$GeneID %in% msl2_in_genes_gene$geneID,"2.MSL2",
ifelse(S2_DEG$GeneID %in% peak_SXL_genes$GeneID,"3.SXL","4.non"))) %>% as.factor()
S2_DEG$DEG <- ifelse(S2_DEG$GeneID %in% S2_up$GeneID ,"2.UP","1.Down") %>% as.factor()
S2_DEG <- S2_DEG[!S2_DEG$S2.TYPE == "4.non",]
test_S2_UP <- S2_DEG[S2_DEG$DEG == "2.UP",]
test_S2_Down <- S2_DEG[S2_DEG$DEG == "1.Down",]
library(ggalluvial)
rownames(S2_DEG) <- S2_DEG$GeneID
S2_DEG <- S2_DEG[,c(-1,-2)]
S2_DEG$ID <- paste0(S2_DEG$S2.TYPE,"_",S2_DEG$DEG)
plot_dat <- as.data.frame(table(S2_DEG$ID))
colnames(plot_dat) <- c("ID","num")
plot_dat$S2.TYPE <- str_split(plot_dat$ID,"\\_",simplify = T)[,1]
plot_dat$DEG <- str_split(plot_dat$ID,"\\_",simplify = T)[,2]
rownames(plot_dat) <- plot_dat$ID
plot_dat <- subset(plot_dat,select=c("S2.TYPE","DEG","num"))
library(ggalluvial)
plot_dat$S2.TYPE <- factor(plot_dat$S2.TYPE,
levels = c("1.SXL.MSL2","2.MSL2","3.SXL"))
df <- to_lodes_form(plot_dat,
key = "x", value = "stratum", id = "alluvium",
axes = 1:2)
col <- c("#A9CCE3","#EC7063","#7D3C98",
"#A9CCE3","#EC7063","#7D3C98",
"#A9CCE3","#EC7063","#7D3C98")
setwd("E:/220625_PC/R workplace/220320_SXL/202404_Fig/241106/")
pdf("sankey_S2_only.pdf",width = 7,height = 6)
ggplot(df, aes(x = x, y=num, fill=stratum, label=stratum,
stratum = stratum, alluvium = alluvium))+
geom_flow(width = 0.22,
curve_type = "arctangent",
alpha = 0.6,
color = "white")+
geom_stratum(width = 0.20, alpha=0.8)+
geom_text(stat = 'stratum', size = 3, color = 'black')+
scale_fill_manual(values = col)+
theme_void()+
theme(legend.position = 'none')
dev.off()
|